Researchers in the United States have conducted a study showing that Moderna’s mRNA-1273 vaccine neutralizes all circulating variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) tested to date.
The novel SARS-CoV-2 virus is the agent responsible for the coronavirus disease 2019 (COVID-19) pandemic that has now caused more than 2.14 million deaths worldwide.
Clinical trial findings have previously shown that Moderna’s mRNA-1273 elicits potent neutralizing activity against SARS-CoV-2 and is highly effective at preventing symptomatic COVID-19 and severe disease.
However, concerns have arisen that the recently emerged SARS-CoV-2 variants in the United Kingdom (B.1.1.7 lineage) and the South African (B.1.351 lineage) could avoid targeting by neutralizing antibodies elicited by natural infection or vaccination.
Now, a study conducted by researchers from Moderna Inc. in Cambridge, Massachusetts, and the National Institute of Allergy and Infectious Diseases in Bethesda, Maryland, has demonstrated the efficacy of mRNA-1273 against all known circulating strains of the virus.
Nevertheless, active viral surveillance and testing of vaccines against emerging variants must continue so that strain-matched vaccines can be developed if warranted, says Darin Edwards and colleagues.
A pre-print version of the research paper is available on the bioRxiv server, while the article undergoes peer review.Study: mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. Image Credit: Kateryna Kon / Shutterstock
The vaccine received Emergency Use Authorization in December 2020
Moderna’s mRNA-1273 vaccine received Emergency Use Authorization from the US Food and Drug Administration on the 18th December last year (2020) after a phase 3 trial demonstrated efficacy of around 94%.
However, the increased transmission rates and higher viral loads associated with the recently emerged B.1.1.7 (UK) and B.1.351 (South Africa) variants have raised concerns that these strains may potentially circumvent immunity conferred by natural infection or vaccination.
The genome of the B.1.1.7 variant contains seventeen mutations. Eight of these mutations are located on a protein called spike – the main structure the virus uses to bind to and infect host cells.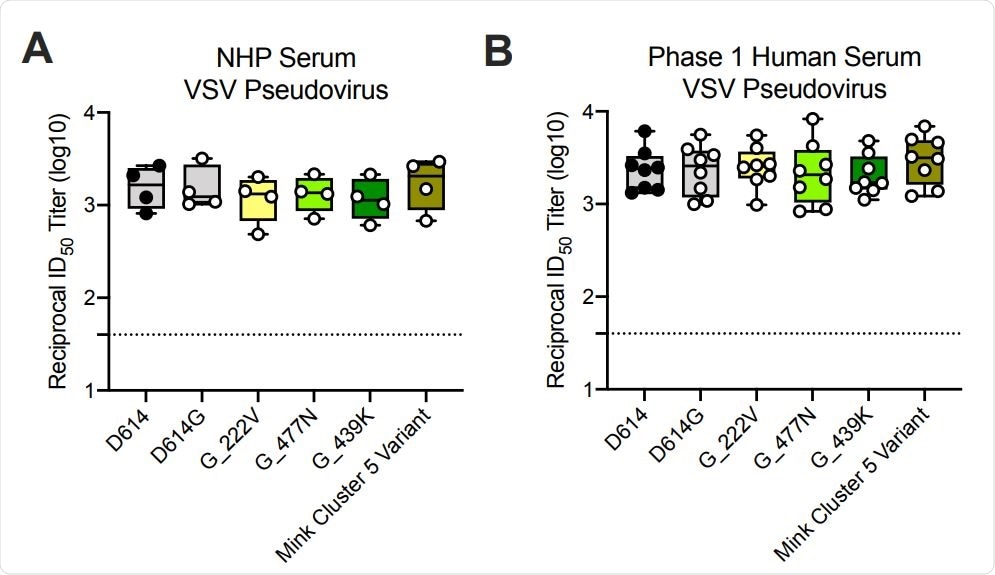 Ability of mRNA-1273 immune sera from NHPs and humans to neutralize SARSCoV-2 pseudoviruses representing early variants. (A) Rhesus macaques (NHPs) were immunized with 30 µg mRNA-1273 on a prime-boost schedule, and sera were collected 4 weeks post-boost. (B) Phase 1 trial participants were immunized with 100 µg mRNA-1273 on a prime-boost schedule, and sera were collected 1 week post-boost. Neutralization was measured by a recombinant VSV-based SARS-CoV-2 pseudovirus neutralization assay incorporating full-length spike protein of the Wuhan isolate (D614) or the indicated spike variants (D614G, A222V-D614G, S477N-D614G, N439K-D614G, mink cluster 5 variant). Min to max box plots, with the box from 25-75% and the median value denoted by the line. The horizontal dotted lines indicate the lower limit of quantification (LLOQ=40). G=D614G.
Ability of mRNA-1273 immune sera from NHPs and humans to neutralize SARSCoV-2 pseudoviruses representing early variants. (A) Rhesus macaques (NHPs) were immunized with 30 µg mRNA-1273 on a prime-boost schedule, and sera were collected 4 weeks post-boost. (B) Phase 1 trial participants were immunized with 100 µg mRNA-1273 on a prime-boost schedule, and sera were collected 1 week post-boost. Neutralization was measured by a recombinant VSV-based SARS-CoV-2 pseudovirus neutralization assay incorporating full-length spike protein of the Wuhan isolate (D614) or the indicated spike variants (D614G, A222V-D614G, S477N-D614G, N439K-D614G, mink cluster 5 variant). Min to max box plots, with the box from 25-75% and the median value denoted by the line. The horizontal dotted lines indicate the lower limit of quantification (LLOQ=40). G=D614G.
A 2021 study has already shown that one of these mutations – the 69-70 deletion – is associated with reduced sensitivity to neutralization by convalescent serum from previously infected individuals.
The spike protein of the B.1.351 variant contains even more mutations than B.1.1.7. Three of these are located on the spike’s receptor-binding domain (RBD), the region that binds to host cells as the initial stage of infection.
“As the RBD is the predominant target for neutralizing antibodies, these mutations could impact the effectiveness of monoclonal antibodies already approved and in advanced development as well as of polyclonal antibody elicited by infection or vaccination in neutralizing the virus,” says Edwards and the team.
Related Stories
- Colchicine may have therapeutic potential against COVID-19
- Viruses like SARS-CoV-2 may be more widespread than previously thought
- Neurological and psychiatric complications six months after COVID-19 recovery
Data from another 2021 study suggested that a key mutation in the B.1.351 variant –E484K – also confers resistance to SARS-CoV-2 neutralizing antibodies.
What did the researchers do?
The researchers assessed the neutralization capacity of sera from eight Phase 1 clinical trial participants and from non-human primates (NHPs) that had received the mRNA-1273 vaccine.
Neutralization was measured against the original SARS-CoV-2 isolate from Wuhan, China; the D614G variant that then became dominant; the B.1.1.7 and B.1.351 variants, and other variants that have previously emerged (20E, 20A.EU2, D614G-N439K, and mink cluster 5 variant).
What did they find?
All samples from both the clinical trial participants and NHPs fully neutralized all of the SARS-CoV-2 strains tested.
No significant impact on neutralization capacity against the B.1.1.7 variant was observed whether antibody titers were measured against the full set of mutations or the 69-70 deletion only.
“Although these mutations have been reported to lessen neutralization from convalescent sera, sera from the Phase 1 participants and NHPs immunized with mRNA-1273 were able to neutralize the B.1.1.7 variant to the same level as the D614G virus,” writes the team.
Of all the variants tested, the only one that reduced levels of neutralizing antibodies was B.1.351. Sera from the clinical trial participants showed a 2.7-fold reduction in neutralization when the three mutations (K417N, E484K, N501Y) found in the spike RBD were included.
Importantly, when the full set of B.1.351 mutations were included, a 6.4-fold reduction in neutralization was observed.
All samples still neutralized the B.1.351 variant
However, neutralizing titers remained above the levels previously found to confer protection in NHPs and all samples from both the clinical trial participants and NHPs fully neutralized the B.1.351 variant. Viral escape was not detected in any of the samples tested.
“These data demonstrate reduced but still significant neutralization against the full B.1.351 variant following mRNA-1273 vaccination,” writes the team.
The researchers say the emergence of strain variants and the ability of the virus to partially overcome natural or vaccine-induced immunity does serve as a call to action and that active viral surveillance and testing of protection against new viral variants must continue.
“The mRNA platform allows rapid design of vaccine antigens that incorporate key mutations,” says Edwards and colleagues.
“Strain-matched vaccines can be developed in response to these variants, either to understand how we might evolve the vaccine-induced immune response through boosting, or to assess the cross-protection provided by a primary series,” concludes the team.
*Important Notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.Journal reference:
- Edwards D, et al. mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-2 variants. bioRxiv, 2021. doi: https://doi.org/10.1101/2021.01.25.427948, https://www.biorxiv.org/content/10.1101/2021.01.25.427948v1
Source: News Medical Life Sciences